AMEBA: Advanced MEtabolic Branchpoint Analysis
AMEBA is the optional addon of metano which allows metabolic branch point analysis and flux visualization. The software is released under the terms of the GNU General Public License version 3.
Use AMEBA online
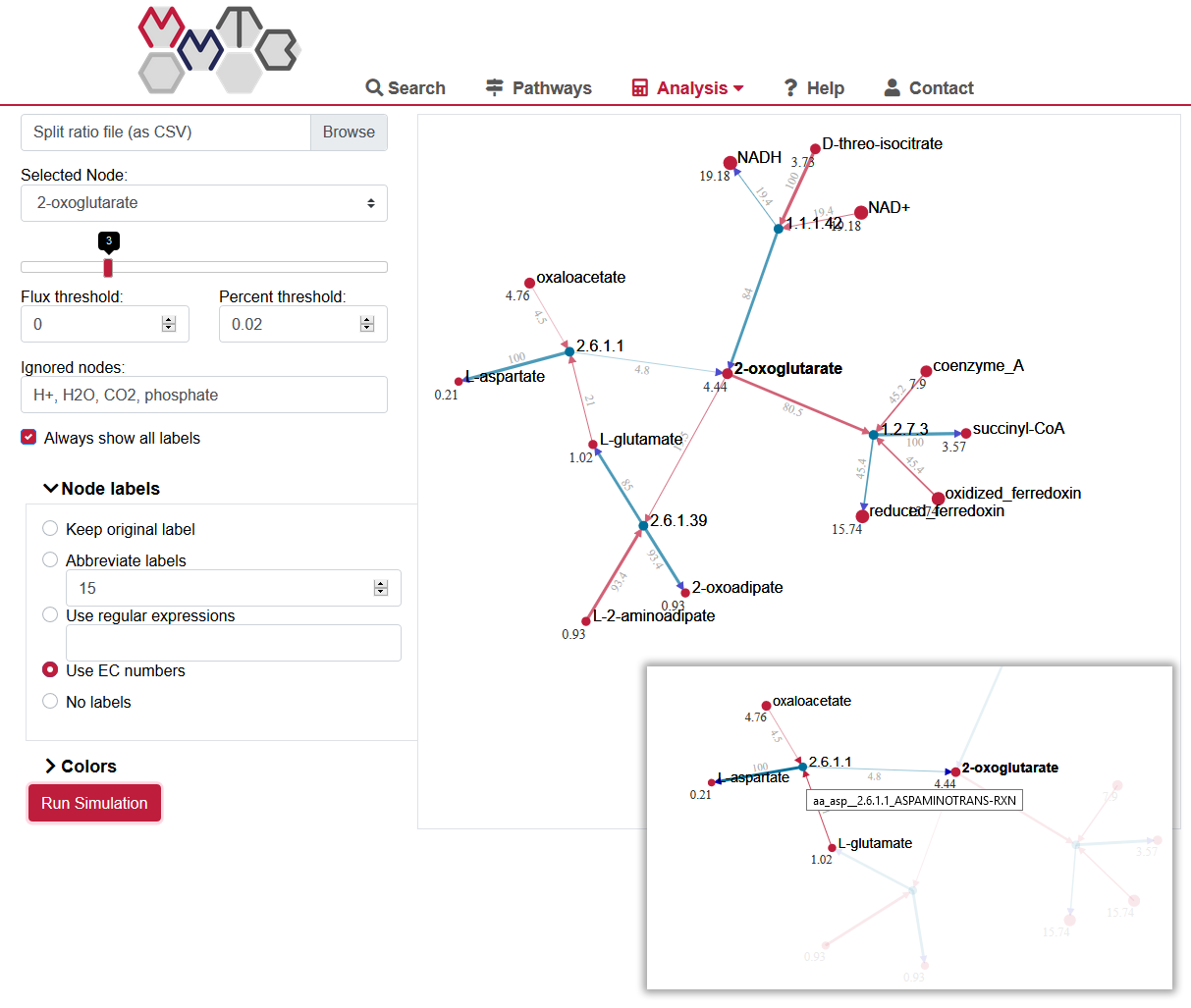
Since AMEBA is also part of MMTB, it is highly recommended to use the online version of split-ratio visualization.
Installation
Please note that AMEBA requires at least Python in version 2.7 to work properly.
Install the following dependencies using the package manager of your distribution:
Under Debian/Ubuntu type:
sudo apt-get install python-gi graphviz-dev python-pygraphviz gir1.2-gtk-3.0 python-gi-cairo
Under openSUSE type:
sudo zypper install python-gobject graphviz-devel gtk3-devel python-cairo
Install AMEBA
Install AMEBA via pip:
sudo pip install ameba
Alternatively you can download AMEBA manually from the PyPI Website and install it by typing:
sudo pip install ameba*.tar.gz
Usage
For an explanation of all command line options type:
ameba -h
A working example can be found in the ameba example folder in the dist-packages location of your python installation. In the case of Ubuntu 12.04 this is:
/usr/local/lib/python2.7/dist-packages/ameba/example/
Alternatively, you can find the examples in the ameba installation folder (under /src/example).
The example folder contains the reaction file (rea_Ecoli_iAF1260_bigg.txt) of the iAF1260 model of
E. coli (Feist et al, 2007) and the corresponding
parameter file for an aerobic
glucose scenario (para_Ecoli_iAF1260_bigg.txt) in the metano format.
Furthermore, we provide a precomputed solution calculated by the FBA
implementation of metano (flux__Ecoli_iAF1260_bigg.txt). The file bpa.ini
contains display options for ameba, which you may want to adjusted to your
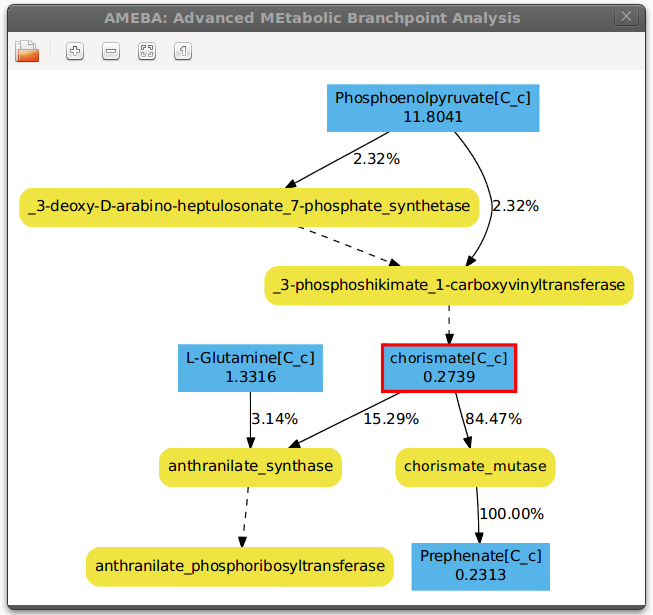
needs. After changing to the example folder AMEBA can be started at the node of the metabolite chorismate by
typing:
ameba -r rea_Ecoli_iAF1260_bigg.txt -s flux__Ecoli_iAF1260_bigg.txt -c bpa.ini -n chorismate[C_c]
Now you can navigate through the metabolic network by clicking on any node in the GUI.
AMEBA was developed by René Rex in the Department of Bioinformatics and Biochemistry at Braunschweig University of Technology.

